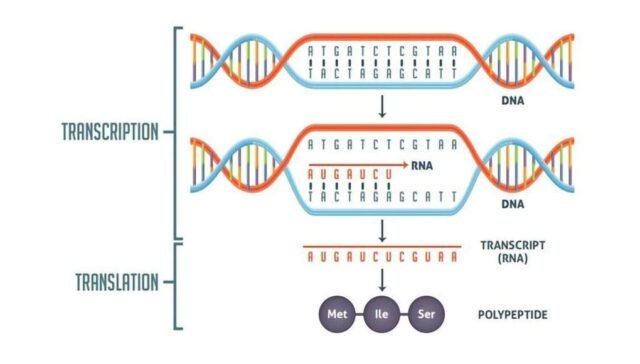
1. What is transcription, and how does it occur in prokaryotic and eukaryotic cells?
Answer:
Transcription is the process by which an RNA molecule is synthesized from a DNA template. In prokaryotic cells, transcription occurs in the cytoplasm because there is no nucleus. RNA polymerase binds to the DNA at the promoter region and synthesizes a complementary RNA strand in the 5′ to 3′ direction. In eukaryotic cells, transcription occurs in the nucleus. Here, RNA polymerase binds to the promoter region of the gene, and after transcription, the pre-mRNA is modified by adding a 5′ cap, splicing, and a 3′ poly-A tail. The mature mRNA then exits the nucleus and enters the cytoplasm for translation.
2. Explain the role of RNA polymerase in the transcription process.
Answer:
RNA polymerase is the enzyme responsible for synthesizing RNA during transcription. It binds to the promoter region of a gene, unwinds the DNA, and synthesizes an RNA strand complementary to the DNA template strand in the 5′ to 3′ direction. RNA polymerase does not require a primer to initiate transcription and continues elongating the RNA strand until it reaches a termination signal. The RNA polymerase then dissociates from the DNA template, releasing the newly formed RNA molecule.
3. Describe the process of RNA splicing in eukaryotic cells.
Answer:
RNA splicing is a post-transcriptional modification process in eukaryotic cells. After the RNA is transcribed, it is called pre-mRNA and contains both exons (coding regions) and introns (non-coding regions). During splicing, the introns are removed, and the exons are joined together by a complex called the spliceosome. This process results in the formation of a mature mRNA molecule that can be exported from the nucleus to the cytoplasm for translation. Splicing is essential because it ensures that only the coding regions (exons) of the gene are used to create proteins.
4. What are codons, and how do they function in translation?
Answer:
Codons are sequences of three nucleotides in mRNA that specify a particular amino acid during protein synthesis. Each codon corresponds to one of the 20 amino acids that make up proteins or a stop signal. For example, the codon “AUG” codes for the amino acid methionine, which also serves as the start signal for translation. The ribosome reads the mRNA codons in sets of three during translation, and each codon pairs with an appropriate tRNA carrying the corresponding amino acid, ultimately forming a polypeptide chain.
5. What is the role of tRNA in the translation process?
Answer:
Transfer RNA (tRNA) is responsible for bringing amino acids to the ribosome during translation. Each tRNA molecule has an anticodon that is complementary to an mRNA codon. The tRNA carries the corresponding amino acid and binds to the mRNA codon at the ribosome. The ribosome facilitates the formation of peptide bonds between the amino acids brought by tRNA, resulting in the elongation of the growing polypeptide chain. Once the tRNA has delivered its amino acid, it exits the ribosome, and another tRNA enters to continue the process.
6. Describe the role of ribosomes in translation.
Answer:
Ribosomes are the molecular machines that facilitate protein synthesis during translation. They consist of two subunits, a large and a small subunit. The small subunit binds to the mRNA, while the large subunit helps catalyze the formation of peptide bonds between amino acids. The ribosome has three sites where tRNAs bind: the A (aminoacyl) site, where the incoming tRNA binds; the P (peptidyl) site, where the tRNA holding the growing polypeptide chain resides; and the E (exit) site, where the tRNA exits after it has delivered its amino acid.
7. What is the start codon, and why is it important in translation?
Answer:
The start codon is the first codon of mRNA that signals the beginning of translation. In eukaryotes and prokaryotes, the start codon is “AUG,” which codes for the amino acid methionine. The presence of this start codon initiates the translation process, as it is recognized by the ribosome, which assembles the appropriate tRNA carrying methionine to begin protein synthesis. The start codon sets the reading frame for translation and ensures that the correct amino acid sequence is formed.
8. What are the differences between prokaryotic and eukaryotic transcription and translation?
Answer:
In prokaryotes, transcription and translation occur simultaneously in the cytoplasm because there is no nucleus. The mRNA is directly translated into protein after being transcribed. In eukaryotes, transcription occurs in the nucleus, where mRNA is synthesized, processed (including splicing, 5′ cap addition, and 3′ poly-A tail addition), and then transported to the cytoplasm for translation. Eukaryotic cells also have more complex regulation of gene expression and RNA processing, while prokaryotic cells have simpler mechanisms.
9. Explain the process of termination in transcription.
Answer:
Termination in transcription occurs when RNA polymerase reaches a specific sequence of DNA known as the terminator. In prokaryotes, the termination sequence causes the RNA polymerase to dissociate from the DNA template, releasing the newly synthesized RNA molecule. In eukaryotes, termination is more complex, often involving the recognition of specific sequences or the cleavage of the newly synthesized RNA followed by polyadenylation (addition of a poly-A tail). This signals the end of transcription and the release of the pre-mRNA.
10. How do mutations in the DNA affect transcription and translation?
Answer:
Mutations in the DNA can affect both transcription and translation. A mutation in the promoter region can prevent RNA polymerase from binding, halting transcription. Mutations in the coding region can alter the mRNA sequence, leading to changes in the amino acid sequence of the resulting protein. For example, a point mutation in the DNA might change a codon, leading to a different amino acid being incorporated into the protein (missense mutation) or causing a premature stop codon (nonsense mutation), which may result in a truncated protein. Frame-shift mutations can also occur if nucleotides are inserted or deleted, shifting the reading frame and causing widespread changes in the protein.
11. What are exons and introns, and how do they relate to mRNA processing?
Answer:
Exons are the coding regions of a gene that contain the instructions for making proteins. Introns are non-coding regions that are transcribed into pre-mRNA but are not part of the final protein. During mRNA processing in eukaryotes, introns are removed, and exons are spliced together to form a continuous mRNA sequence. This process ensures that only the coding regions are included in the mature mRNA, which can then be translated into protein.
12. What are the different types of RNA involved in protein synthesis?
Answer:
There are three main types of RNA involved in protein synthesis:
- mRNA (messenger RNA): Carries the genetic information from the DNA in the nucleus to the ribosome, where it is translated into protein.
- tRNA (transfer RNA): Brings amino acids to the ribosome during translation, matching the mRNA codons with the appropriate amino acids.
- rRNA (ribosomal RNA): Forms the core structural and catalytic components of the ribosome, aiding in the assembly of amino acids into a protein.
13. What is the role of the 5′ cap and 3′ poly-A tail in mRNA processing?
Answer:
The 5′ cap and 3′ poly-A tail are modifications made to the pre-mRNA molecule in eukaryotes. The 5′ cap, added to the beginning of the mRNA, helps protect the mRNA from degradation, aids in its export from the nucleus, and helps the ribosome recognize the mRNA for translation. The 3′ poly-A tail, added to the end of the mRNA, also protects the mRNA from degradation and plays a role in mRNA stability and translation efficiency. These modifications are essential for the proper function of mRNA in protein synthesis.
14. What is the process of elongation in translation?
Answer:
Elongation in translation refers to the step-by-step addition of amino acids to the growing polypeptide chain. After the ribosome binds to the mRNA and the start codon is recognized, tRNA molecules carry amino acids to the ribosome, where they are added to the growing chain. Each tRNA recognizes a codon in the mRNA and binds to it through complementary base pairing between the tRNA anticodon and the mRNA codon. As the ribosome moves along the mRNA, new amino acids are added to the chain, and the process continues until a stop codon is reached.
15. What is a stop codon, and what happens when it is encountered during translation?
Answer:
A stop codon is a sequence of three nucleotides (UAA, UAG, or UGA) in mRNA that signals the end of the translation process. When the ribosome encounters a stop codon, it does not bind any tRNA carrying an amino acid. Instead, release factors bind to the stop codon, prompting the ribosome to release the newly synthesized polypeptide chain and dissociate from the mRNA. This marks the termination of translation, and the protein is released to undergo folding and other modifications.
16. How does the ribosome ensure the accuracy of translation?
Answer:
The ribosome ensures the accuracy of translation through a combination of mechanisms. First, the tRNA anticodon is complementary to the mRNA codon, ensuring that the correct amino acid is incorporated. Second, the ribosome has a proofreading function that checks for proper codon-anticodon pairing. If the pairing is incorrect, the tRNA is rejected, and the process is repeated with a correct tRNA. Additionally, the ribosome catalyzes the formation of peptide bonds only when the correct tRNA is in place, ensuring that the polypeptide chain is synthesized accurately.
17. What is the function of the large ribosomal subunit in translation?
Answer:
The large ribosomal subunit is responsible for the formation of peptide bonds between amino acids during translation. Once the small ribosomal subunit binds to the mRNA and initiates translation, the large subunit attaches to form a functional ribosome. The large subunit contains the enzymatic activity necessary to catalyze the formation of peptide bonds between adjacent amino acids brought by tRNAs. This process is essential for the elongation of the polypeptide chain during translation.
18. How is the translation process terminated?
Answer:
Translation is terminated when a stop codon (UAA, UAG, or UGA) is encountered by the ribosome. The stop codon does not correspond to any amino acid, so no tRNA binds to it. Instead, release factors bind to the ribosome and trigger the release of the newly synthesized polypeptide chain. The ribosome then dissociates from the mRNA, and the translation complex disassembles, marking the end of the translation process. The protein is then released to undergo further folding and modifications.
19. What is the role of the promoter in transcription?
Answer:
The promoter is a specific sequence of DNA that marks the start site for transcription. It is located upstream of the gene to be transcribed. In prokaryotes, the RNA polymerase binds directly to the promoter to initiate transcription. In eukaryotes, additional transcription factors are required to bind to the promoter before RNA polymerase can begin transcription. The promoter ensures that transcription begins at the correct location and is crucial for the regulation of gene expression.
20. What is the significance of the genetic code in translation?
Answer:
The genetic code is a set of rules that determines how the sequence of nucleotides in DNA and RNA is translated into an amino acid sequence in proteins. The genetic code is universal across most organisms, ensuring consistency in protein synthesis. Each triplet of nucleotides (codon) in mRNA corresponds to one specific amino acid. The code also includes stop codons, which signal the end of translation. This ensures that the correct protein is synthesized according to the instructions encoded in the DNA.
21. What is the role of the transcription factors in eukaryotic transcription?
Answer:
Transcription factors are proteins that help regulate gene expression in eukaryotic cells. They bind to specific sequences in the promoter region of the gene and either promote or inhibit the binding of RNA polymerase. Transcription factors can activate or repress transcription by interacting with other regulatory proteins, such as co-activators or co-repressors, and by modifying the chromatin structure to make the gene more or less accessible for transcription. This regulation allows cells to control which genes are expressed at any given time.
22. Explain the role of enhancers and silencers in transcription regulation.
Answer:
Enhancers and silencers are regulatory DNA sequences that influence the transcription of genes. Enhancers are regions of DNA that increase the rate of transcription when bound by specific transcription factors. They can be located far from the gene they regulate and may function by bending the DNA to bring the enhancer closer to the promoter. Silencers, on the other hand, are sequences that repress transcription when bound by repressive transcription factors. Both enhancers and silencers provide fine-tuned control over gene expression.
23. What is the role of the spliceosome in RNA splicing?
Answer:
The spliceosome is a complex of RNA and protein molecules responsible for the removal of introns and the joining of exons during RNA splicing in eukaryotes. It consists of several small nuclear RNAs (snRNAs) and associated proteins that work together to recognize splice sites, cut out the introns, and join the exons. This process ensures that the final mRNA transcript contains only the coding sequences and is ready for translation in the cytoplasm.
24. How does alternative splicing contribute to genetic diversity?
Answer:
Alternative splicing is a process in which different combinations of exons are joined together, allowing a single gene to produce multiple protein isoforms. This increases the diversity of proteins that can be produced from a single gene and allows the organism to generate a wide range of functional proteins. Alternative splicing is regulated by various factors, including spliceosome components and regulatory proteins, and plays a crucial role in tissue-specific gene expression and developmental processes.
25. What are the differences between the three types of RNA (mRNA, tRNA, and rRNA)?
Answer:
- mRNA (messenger RNA): Carries the genetic instructions from DNA to the ribosome for protein synthesis. It is transcribed from the DNA template and contains the codons that dictate the amino acid sequence in a protein.
- tRNA (transfer RNA): Delivers amino acids to the ribosome during translation. Each tRNA has an anticodon that pairs with a corresponding mRNA codon, ensuring the correct amino acid is incorporated into the polypeptide chain.
- rRNA (ribosomal RNA): Forms the structural and catalytic core of the ribosome. It helps align the mRNA and tRNAs and catalyzes the formation of peptide bonds between amino acids during translation.
26. How is the fidelity of transcription ensured?
Answer:
The fidelity of transcription is ensured by several mechanisms. RNA polymerase has proofreading activity, where it can detect and correct errors in the RNA strand as it is synthesized. Additionally, the process of transcription involves several checkpoints and regulatory proteins that help minimize mistakes in the RNA sequence. In eukaryotes, mRNA undergoes additional processing, including splicing, that further ensures the final transcript is accurate before being translated.
27. What is the role of the 5′ cap in eukaryotic mRNA processing?
Answer:
The 5′ cap is added to the beginning of eukaryotic mRNA molecules shortly after transcription begins. It serves several purposes: it protects the mRNA from degradation by exonucleases, facilitates the export of the mRNA from the nucleus to the cytoplasm, and helps in the recognition of the mRNA by the ribosome during translation. The 5′ cap also plays a role in the stability of the mRNA molecule and its translation efficiency.
28. How does the ribosome facilitate translation initiation?
Answer:
The ribosome facilitates translation initiation by assembling on the mRNA at the start codon. The small ribosomal subunit binds to the 5′ cap and the mRNA sequence, locating the start codon. Once the correct codon (AUG) is found, the large ribosomal subunit attaches to the small subunit, and translation begins. The initiator tRNA carrying methionine binds to the start codon, and the ribosome begins synthesizing the protein by elongating the polypeptide chain.
29. What are the key steps in the elongation cycle of translation?
Answer:
The elongation cycle of translation consists of three main steps:
- Aminoacyl tRNA binding: The ribosome binds a new tRNA carrying an amino acid to the A site based on complementary base pairing between the tRNA anticodon and the mRNA codon.
- Peptide bond formation: The ribosome catalyzes the formation of a peptide bond between the amino acid in the A site and the growing polypeptide chain in the P site.
- Translocation: The ribosome moves along the mRNA by one codon, shifting the tRNA from the A site to the P site and moving the empty tRNA to the E site, where it exits the ribosome.
30. What is the significance of the genetic code being redundant yet unambiguous?
Answer:
The genetic code is redundant because multiple codons can specify the same amino acid. For example, the codons UUU and UUC both code for phenylalanine. This redundancy helps minimize the impact of mutations, as a change in the third nucleotide of a codon may still result in the same amino acid being incorporated into the protein. However, the code is unambiguous because each codon specifies only one amino acid, ensuring that the correct protein is synthesized according to the genetic instructions.
These questions and answers cover a wide range of concepts related to the processes of transcription and translation.